AMDX
AmoyDx® Essential NGS Panel | 8.06.27401X024I
- SKU:
- AMDX-AM-8.06.27401X024I-GEN
- Availability:
- IN STOCK
Description
AmoyDx® Essential NGS Panel | 8.06.27401X024I | Gentaur UK, US & Europe Distribution
Intended Use
The AmoyDx® Essential NGS Panel is a next-generation sequencing (NGS) based in vitro diagnostic assay intended for qualitative detection of single nucleotide variants (SNVs), insertions and deletions (InDels), gene fusions, and copy number variations (CNVs) in 10 oncogenic driver genes, using DNA isolated from formalin-fixed paraffin embedded (FFPE) tumour tissue specimens, or circulating cell-free DNA (cfDNA) isolated from plasma derived from anti-coagulated peripheral whole blood specimens. The detection of CNVs is available for tissue-derived DNA only. The assay is intended to provide tumor mutation profiling to be used by qualified health care professionals in accordance with professional guidelines in oncology for patients with non-small cell lung cancer (NSCLC) and colorectal cancer (CRC).
This assay is not automated and is for laboratory professional use only.
Principles of the Procedure
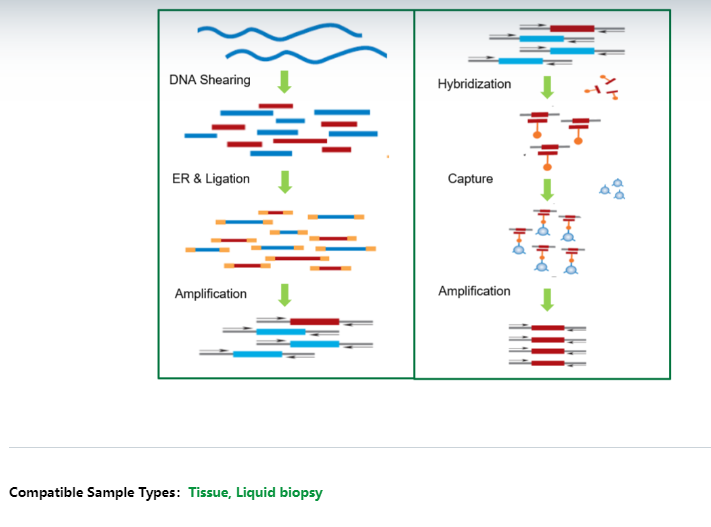
The test kit is based on dual-directional capture (ddCAP) technology which is a targeted next generation sequencing method that uses biotinylated oligonucleotide baits (probes) to hybridize to the target regions. The test kit is designed for use with fragmented genomic DNA (gDNA) or cfDNA. During the library construction process, each individual DNA molecule is tagged with a unique molecular index (UMI) at both ends, which allows high sensitivity in variant detection by eliminating any library amplification and sequencing bias.
For FFPE tissue samples, the extracted DNA should be sheared into short fragments, using either mechanical methods (e.g.,ultrasonication shearing) or enzymatic digestion, then the purified fragmented DNA can be used for downstream library preparation. For plasma samples, the extracted cfDNA can be used directly to downstream library preparation.
The test kit includes the reagents and enzymes needed for library preparation. First, the fragmented DNA or cfDNA are incubated with end repair enzyme and reagents to get the blunt-ended fragments with dA-tails, then the DNA fragments are ligated to adaptors with complementary dT-overhangs, then the adaptor-ligated DNA fragments are size-selected through AMPure beads, then the PCR amplification is performed to enrich the libraries and each library is tagged with unique dual index. Next, the library is performed with target enrichment, the process including denature the double-strand library, hybridize biotinylated probes to the complementary target DNA, and enrich the captured target DNA using streptavidin beads. Finally, the universal PCR amplification is performed to enrich the target libraries.
After quality control (QC), the qualified libraries could be sequenced on Illumina sequencing platform. The sequencing data can be analyzed by AmoyDx NGS data analysis system (ANDAS) to detect the genomic variants in the target region.
Testing Procedure
Additional Information
Size: |
24 Tests/Kit |
Status: |
CE marked |
Type: |
Bulk |
PCR Instrument (stated in IFU): |
Illumina NextSeq 500, MiSeq |










